The Open Pediatric Brain Tumor Atlas (OpenPBTA) was published in Cell Genomics in May 2023.
The Open Pediatric Brain Tumor Atlas (OpenPBTA) is a global open science initiative, which crowdsources the expertise of multidisciplinary data scientists and pediatric brain cancer experts to gain a deeper understanding of the leading cause of death by disease in children and young adults. This project was developed in response to the prevalence of childhood brain tumors, their low survival rates, and the long-standing challenges that researchers have faced to improve outcomes. Through OpenPBTA, investigators openly analyzed more than 1,000 pediatric brain tumors, while making the results available in real time on GitHub. The project’s manuscript was openly and collaboratively written on GitHub.
Why pediatric brain tumors?
Brain tumors remain the leading cause of cancer-related death in children and young adults. Moreso than many other types of tumors, brain tumors often leave the patient with permanent life-altering physical, cognitive, and psychological effects. It is estimated that 5,900 new cases of pediatric brain tumors will be diagnosed in just the United States in 2022. (Source)
Experts have long faced barriers that have hindered the progress of pediatric brain tumor research. Increasing opportunities for collaboration and improving data sharing practices are critical steps towards tackling these obstacles. OpenPBTA seeks to do just that by bringing together experts with diverse knowledge and skills to collect, analyze, and ultimately molecularly classify pediatric brain tumors. The classification of these tumors can lead to improved analyses and a better understanding of how to treat them.
At Alex's Lemonade Stand Foundation (ALSF), we believe that openly sharing data and resources is the foundation of progress in science and allows researchers to build upon the work of one another more quickly. Often, it can take years for a study to be completed and for the results of an analysis to become public through the peer review and publishing process. With OpenPBTA, analyses and manuscript writing are both conducted publicly, in real time. The data and the analysis code are made openly available as each analysis passes the review process. This rapid dissemination makes it possible for researchers anywhere to gain access to the results of analyses much faster, while promoting transparency and allowing others to participate in the project.
Crowdsourcing expertise
In recent years, cooperative efforts have accelerated the discovery of novel therapies for these difficult-to-treat diseases. Organizations such as Children’s Brain Tumor Network (CBTN) and the Pacific Pediatric Neuro-Oncology Consortium (PNOC) came together to foster collaboration and scientific resource-sharing. In 2018, they jointly launched the Pediatric Brain Tumor Atlas (PBTA), a large collection of pediatric brain tumor data made available through the Gabriella Miller Kids First Portal. Releasing this data opened up an exciting new opportunity for the pediatric brain tumor research community to comprehensively explore and classify the genomic landscapes of these tumors and identify key insights that can inform future treatments.
ALSF’s Childhood Cancer Data Lab (Data Lab), and the Center for Data-Driven Discovery in Biomedicine at Children’s Hospital of Philadelphia leveraged an open contribution model that maximizes the impact of the available PBTA data. We organized and continue to maintain OpenPBTA, which allows researchers from across the world to contribute their expertise to the analysis of this immense collection of tumor samples. To date, 46 collaborators across 18 institutions have contributed to OpenPBTA.
So how does this work?
The success of the project’s open contribution model is made possible by tools that support collaboration and practices that ensure scientific accuracy and reproducibility.
To collectively author the manuscript, collaborators work with a unique software that stores the document in a public repository. This makes it possible for multiple authors to write and review, while saving each version of the manuscript as it is edited and built upon.
Outside parties interested in contributing analyses to OpenPBTA are able to contact project organizers through the project's analysis repository. Here, they can submit a description of their proposed analysis, including their goals and specific plans. Then project organizers and other contributors provide their initial feedback. Before a contribution is accepted as part of the project, the potential contributor must formally request to add their analytical code to the repository. Any such request is subject to rigorous peer review by organizers and maintainers. (Figure 1)
Peer review of code is a unique part of OpenPBTA. Large collaborations like this don’t always require such an involved review process. But to ensure reproducible results, peer review is critical. Code review ensures that more than one person has successfully interpreted and run that code, making it far more likely that others who want to use OpenPBTA’s reproducible workflows for their own research projects can do so.
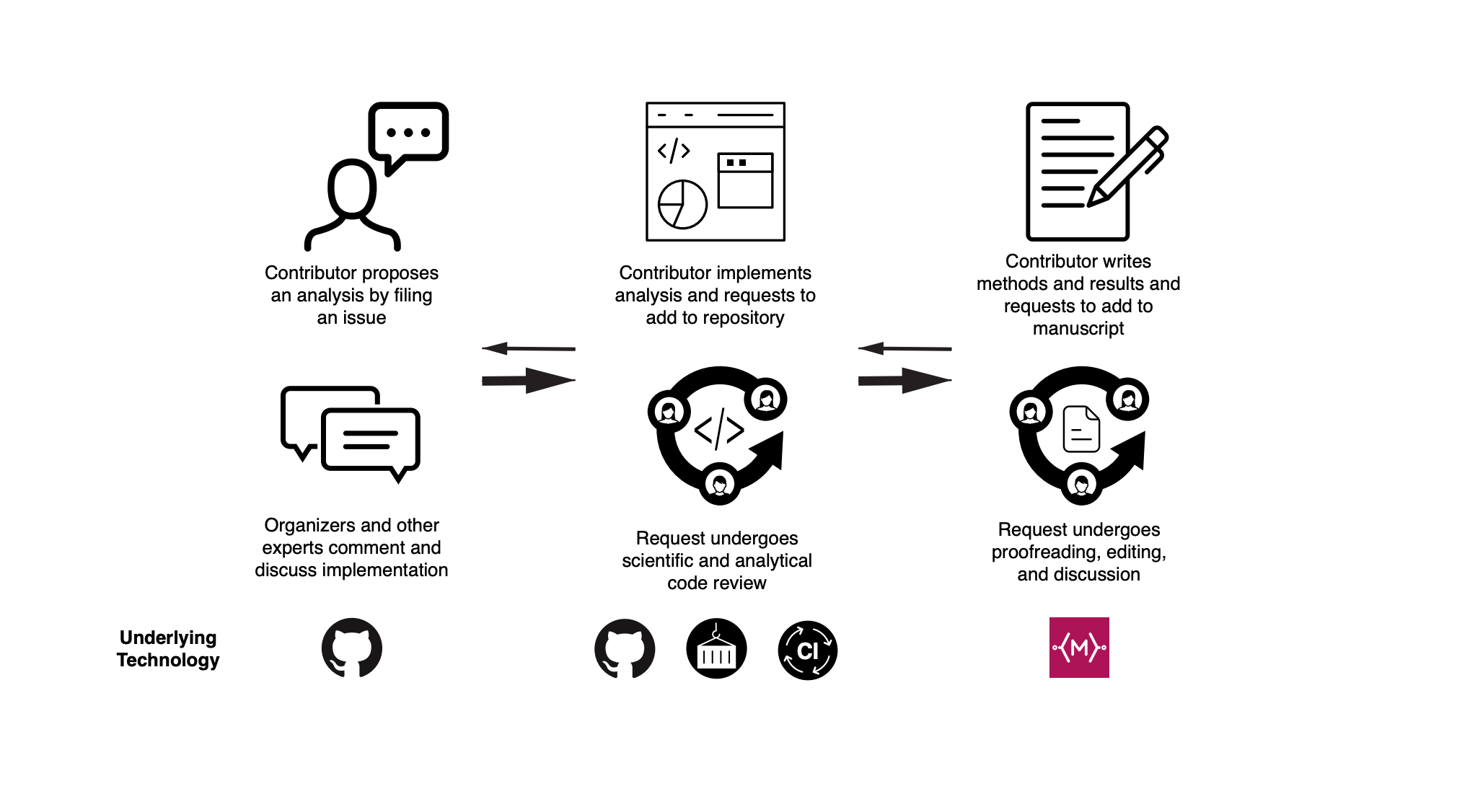
Figure 1: This figure from the OpenPBTA manuscript illustrates how the open contribution model works from the time an interested contributor proposes an analysis.
The results of a shared effort
To our knowledge, OpenPBTA is the largest open effort to analyze a collection of pediatric brain tumor data this size, while collaboratively authoring a manuscript. Over the course of this project, more than 1,000 pediatric brain tumors have been analyzed and all code and processed data is currently available to researchers via GitHub, Cavatica, and PedcBioPortal.
The cooperative analysis of this massive collection also helped vastly improve the data itself. Much of the data was first generated prior to the guidelines described in 2016 by the World Health Organization (WHO) to classify cancers. Following those guidelines, OpenPBTA researchers updated the diagnoses of more than half of the tumor samples in this collection. These updates and improvements ensure that future research performed using PBTA data is more reliable and more robust.
The collaborative nature of this project further provides researchers with ample opportunity to share information, improve their skill sets, and learn from one another. Collaborators cultivate skills and best practices for robust and reproducible analysis and gain knowledge they can apply to their own research projects. The reproducible workflows developed through OpenPBTA are already being used by others to study pediatric brain tumor data, as well as other childhood cancers.
What’s next?
The OpenPBTA manuscript has now been submitted for peer-reviewed publication. Read the latest version of it here! Organizers and maintainers look forward to sharing the pivotal results of this project with the research community. They hope to see more pediatric cancer research projects utilizing OpenPBTA’s reproducible workflows and adapting collaborative, open contribution models.
Innovative projects like OpenPBTA will accelerate the discovery of more cures and better treatments for hard-to-treat diseases like pediatric brain tumors. Support the Data Lab’s projects and their efforts to foster collaboration in the scientific community.
1. Some examples of this include the National Cancer Institute’s (NCI) Molecular Targets Platform and the Open Pediatric Cancer (OpenPedCan) project at the Children’s Hospital of Philadelphia.

